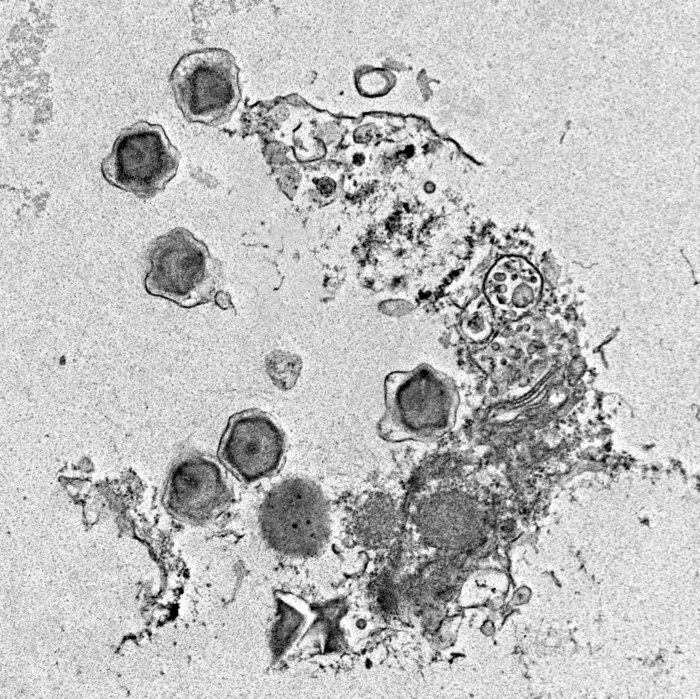
In a groundbreaking study set to redefine our understanding of oceanic ecosystems, researchers from the University of Miami’s Rosenstiel School of Marine, Atmospheric, and Earth Science have uncovered an astonishing diversity of giant viruses that wield significant influence over marine life and environmental health. These colossal entities, often overlooked due to their immense size and complex genomes, are now recognized as pivotal agents in shaping the survival and metabolic activities of protists—single-celled marine microorganisms fundamental to the ocean’s food web. The team’s findings, published in the prestigious journal Nature npj Viruses, document the discovery of 230 previously unknown giant virus genomes, illuminating their intricate roles in oceanic biogeochemistry and offering novel insights into viral manipulation of host cellular processes.
Giant viruses inhabit a unique niche in marine environments. Unlike typical viruses, their genomes encode proteins commonly associated with cellular organisms, empowering them to intricately reprogram their protist hosts during infection. This relationship is critical, as protists—including various algae species—form the foundation of marine food chains and significantly influence global carbon cycling. The newly identified viruses were unearthed through sophisticated bioinformatic analysis of vast metagenomic datasets collected from diverse oceanic regions, ranging from polar seas to equatorial waters. This expansive sampling underpins the universality and ecological significance of these viral populations.
At the core of this discovery lies an innovative bioinformatics software named BEREN (Bioinformatic tool for Eukaryotic virus Recovery from Environmental metagenomes), developed expressly to detect giant viruses within colossal environmental DNA libraries. Traditional computational pipelines have struggled to accurately identify and classify these viruses because of their complex genetic architectures and the scarcity of reference genomes. BEREN’s design overcomes these limitations by integrating advanced genome recovery and annotation protocols, enabling researchers to reconstruct hundreds of viral genomes from raw sequencing data efficiently. This revolutionary tool not only expands the catalog of known giant viruses but also streamlines future viral ecology studies.
Among the 230 novel genomes characterized in this study, the researchers identified over 530 new functional proteins, including several implicated directly in photosynthesis—a biological process traditionally reserved for plants and certain microbes. The presence of photosynthesis-related genes within viral genomes challenges the conventional understanding of viral function and suggests that giant viruses may actively manipulate their host’s photosynthetic mechanisms to optimize viral replication and survival. Such viral hijacking of host metabolism could have far-reaching implications for carbon fixation and energy flow within marine ecosystems.
The implications of this discovery extend beyond fundamental biology. Harmful algal blooms (HABs), a phenomenon often linked to the proliferation of specific protist species, pose severe health hazards to coastal populations worldwide. These blooms deplete oxygen in marine waters and release toxins that can contaminate seafood and threaten human health. By elucidating the diversity and functional capacity of giant viruses that infect bloom-forming algae, this research provides a new lens through which to predict, monitor, and potentially manage HAB occurrences. Understanding the viral triggers and regulators of protist population dynamics could empower environmental agencies with novel tools for coastal risk mitigation.
This study’s success was made possible by leveraging the computational might of the University of Miami’s Pegasus supercomputer housed at the Frost Institute for Data Science and Computing. The supercomputer’s high-throughput processing capabilities enabled the assembly and analysis of metagenomic datasets often exceeding one gigabase in size, a technical feat that would have been impractical with standard laboratory computing resources. This intersection of marine biology and high-performance computing exemplifies the future of ecological research, where data-driven methodologies uncover hidden patterns within complex natural systems.
Lead author Benjamin Minch emphasizes the remarkable metabolic sophistication encoded within these giant viruses. “Our analysis reveals that giant viruses harbor genes traditionally thought exclusive to cellular life, such as those involved in carbon metabolism and photosynthesis,” Minch remarks. This metabolic toolkit allows viruses not only to reproduce within their hosts but also to commandeer and redirect host metabolic pathways, effectively turning protists into viral factories optimized for efficient replication. Such intricate manipulation highlights the evolutionary innovation of giant viruses and their underestimated role in regulating ocean nutrient cycles.
Co-author Mohammad Moniruzzaman elaborates on the study’s broader significance, noting that deeper insight into giant virus ecology enhances our capacity to foresee environmental changes. “By unraveling the genomic and functional diversity of these viruses, we aim to improve surveillance of harmful algal blooms and better safeguard public health along vulnerable coastlines,” he says. Additionally, some of the newly discovered viral functions may harbor biotechnological potential, with novel enzymes possibly applicable in industrial or pharmaceutical contexts. The dual ecological and applied implications underscore giant viruses as a frontier for multidisciplinary scientific exploration.
A critical advance presented by this research is the concept that viruses are active players in marine biogeochemistry rather than passive parasites. The presence of viral genes modulating carbon metabolism suggests that viral infections contribute to the transformation of organic and inorganic carbon in ocean waters, influencing global carbon cycles and potentially climate regulation. Such viral-host interactions likely affect nutrient availability and oxygen production, with cascading impacts on marine food webs and ecosystem resilience amid environmental change.
The study’s methodology involved a comprehensive meta-analysis of public marine metagenomic data from nine expansive global ocean sampling projects. These datasets, publicly accessible and remarkably diverse, provided a fertile ground for BEREN to mine viral genomes and annotate their functional potential. Comparative genomics against existing giant virus sequences identified unprecedented gene variants and functional modules, expanding the taxonomic and ecological landscape of viral influence in marine microbiology. This systematic approach sets a precedent for leveraging open-access data repositories in virology and environmental sciences.
The discovery of these giant viruses also challenges long-standing notions surrounding virus-host dynamics, urging scientists to reconsider the evolution and classification of viruses. With genomes substantially larger than many bacteria, giant viruses blur the boundaries between viral and cellular life, potentially representing a distinct lineage with complex evolutionary histories. Their ability to incorporate sophisticated metabolic genes points to ancient gene exchanges between viruses and eukaryotic hosts, enriching our understanding of how marine microbial communities have co-evolved over millennia.
Beyond the ecological ramifications, the study underscores a pressing need for novel monitoring strategies that incorporate viral components into existing environmental health frameworks. Traditional water quality assessments rarely account for viral diversity or activity, possibly overlooking key determinants of ecosystem function and pathogen emergence. Integrating bioinformatic tools like BEREN into routine surveillance could revolutionize environmental monitoring by enabling early detection of viral agents implicated in harmful blooms, pollution indicators, and emergent pathogenic threats.
Published on April 21, 2025, this landmark study represents a paradigm shift in marine microbial ecology and opens exciting avenues for future research. The comprehensive expansion of known giant virus diversity catalyzes further inquiry into viral ecology, evolutionary biology, and applied biotechnology. As the scientific community embraces viral contributions to ocean health and biogeochemical cycles, innovative collaborations spanning computational sciences, marine biology, and environmental health will become indispensable in confronting global environmental challenges and harnessing viral capabilities for sustainable solutions.
Subject of Research: Cells
Article Title: Expansion of the genomic and functional diversity of global ocean giant viruses
News Publication Date: 21-Apr-2025
References: https://www.nature.com/articles/s44298-025-00122-z
Image Credits: Grieg Steward, Ph.D. University of Hawai’i at Manoa.
Keywords: Plankton, Biochemical processes, Toxicology, Cell biology, Computational biology, Microbiology