Plant diseases pose significant challenges to agricultural productivity, presenting formidable hurdles that require urgent attention. Left unchecked, these diseases can spread rapidly, inflicting widespread damage on crops and leading to reduced yields and substantial economic losses. Therefore, accurately identifying the pathogens responsible for these diseases is crucial. This identification allows for targeted interventions that minimize risks and effectively mitigate the agricultural impacts.

Credit: Masahiro Nakajima from Tokyo University of Science
Plant diseases pose significant challenges to agricultural productivity, presenting formidable hurdles that require urgent attention. Left unchecked, these diseases can spread rapidly, inflicting widespread damage on crops and leading to reduced yields and substantial economic losses. Therefore, accurately identifying the pathogens responsible for these diseases is crucial. This identification allows for targeted interventions that minimize risks and effectively mitigate the agricultural impacts.
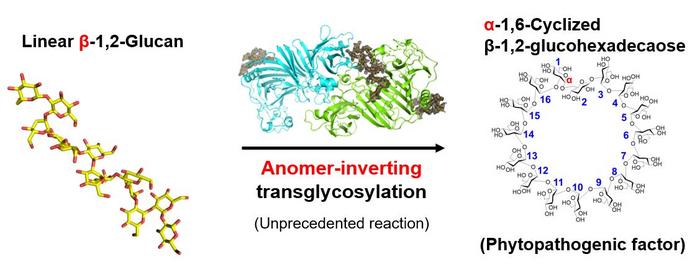
Xanthomonas species are notorious plant pathogens that affect a broad spectrum of hosts, including key crops like rice, wheat, and tomatoes. These pathogens augment their pathogenicity by utilizing α-1,6-cyclized β-1,2-glucohexadecaose (CβG16α) to suppress essential plant defense mechanisms, such as the expression of pathogenesis-related proteins and the accumulation of callose.
In a recent breakthrough published on June 19, 2024, in the Journal of the American Chemical Society, a team of researchers led by Associate Professor Masahiro Nakajima from Tokyo University of Science unveiled a significant discovery. They identified XccOpgD, a glycoside hydrolase (GH186) found in X. campestris pv campestris which plays a pivotal role in the biosynthesis of CβG16α. The research team also included Mr. Sei Motouchi from Tokyo University of Science, Principal Scientist Shiro Komba from the Institute of Food Research, NARO, and Hiroyuki Nakai from Niigata University.
“Glycan structures are intricate and multifaceted and fulfill diverse crucial roles in nature and organisms. Enzymes synthesize and degrade glycans, exhibiting diverse structures and functions that correspond to the glycan diversity. However, our understanding of these enzymes is still limited, which drives the search for new enzymes with varied new potentials,” explains Prof. Nakajima, elaborating on the study’s rationale.
The team conducted biochemical analysis to elucidate the role of XccOpgD in CβG16α biosynthesis. Advanced techniques such as X-ray crystallography were employed as structural analysis to unravel the enzyme’s catalytic mechanism and substrate specificity.
These efforts have yielded profound insights. XccOpgD belongs to the GH186 family, essential for regulating bacterial cell wall components. Unlike the first identified GH186 enzymes, XccOpgD exhibits an unprecedented enzymatic mechanism known as anomer-inverting transglycosylation.
“Reactions of typical GH enzymes are classified into four types by combination of retaining or inverting, and reaction with water (hydrolysis) or sugar (transglycosylation) theoretically. However, one classification is missing somehow in a long history of researches on carbohydrate associated enzymes and we discovered the missing classification. This breakthrough was made possible by unique structural environment, opening new possibilities for enzyme-based glycosylation,” explains Prof. Nakajima. Moreover, the sugar chains synthesized through this mechanism are not merely minor components but rather essential structures utilized by various Gram-negative bacteria in nature for pathogenic purposes.
Detailed studies revealed that linear β-1,2-glucan was converted to cyclic compound and the compound was identified as CβG16α using nuclear magnetic resonance. Structural analysis of the Michaelis complex identified crucial substrate binding residues, further elucidating specific interactions along the glucan chain. Notably, XccOpgD utilizes an anomer-inverting transglycosylation mechanism, with D379 and D291 playing pivotal roles as catalysts.
These findings deepen our understanding and open avenues for developing targeted strategies against Xanthomonas-induced plant diseases. “We are expecting a pesticide concept targeting this enzyme homolog in the future. Unlike fungicides that promote the emergence of drug-resistant bacteria in soil, targeting this enzyme could potentially inhibit pathogenicity without causing sterilization. Enzyme homologs identified in this study may serve as promising structure-based drug targets, offering a potential solution to the issue of drug-resistant bacteria,” says a hopeful Prof. Nakajima.
The discovery of XccOpgD and its role in CβG16α biosynthesis marks a major breakthrough in agriculture. It promises enhanced resilience and food security while mitigating environmental impacts linked to conventional pesticides. Overall, this advancement offers sustainable solutions to global agricultural challenges, promoting environmental stewardship and economic viability for farmers worldwide.
***
Reference
DOI: 10.1021/jacs.4c02579
Authors: Sei Motouchi1, Shiro Komba2, Hiroyuki Nakai3, and Masahiro Nakajima1
Affiliations:
1Department of Applied Biological Science, Tokyo University of Science
2Institute of Food Research, National Agriculture and Food Research Organization
3Faculty of Agriculture, Niigata University
Media contact
Yoshimasa Iwasaki
Public Relations Division
Tokyo University of Science
Email: mediaoffice@admin.tus.ac.jp
Institute of Food Research
National Agriculture and Food Research Organization
Email: www@naro.affrc.go.jp
Public Relations office
Niigata University
Email: pr-office@adm.niigata-u.ac.jp
Funding information
This work was supported in part by JST SPRING (Grant 386 Number JPMJSP2151).
Journal
Journal of the American Chemical Society
Method of Research
Experimental study
Subject of Research
Not applicable
Article Title
Discovery of Anomer-Inverting Transglycosylase: Cyclic Glucohexadecaose-Producing Enzyme from Xanthomonas, a Phytopathogen
Article Publication Date
19-Jun-2024
COI Statement
The authors declare no competing financial interest.