A breakthrough in microbial genomics has emerged with the introduction of BGI’s latest sequencing platform, CycloneSEQ™. This innovative technology, utilizing novel nanopore sequencing techniques, promises to revolutionize the way researchers approach the sequencing of complete bacterial genomes. Following its official launch, an independent benchmarking study has become available, demonstrating its capabilities in producing high-quality genomic data that rivals existing sequencing methodologies. The results not only highlight the strengths of CycloneSEQ™ but also provide a comprehensive dataset that can be utilized by the scientific community for further research and verification of its long-read sequencing approach.
CycloneSEQ™ stands out for its reliance on long-read sequencing, which allows for the direct reading of DNA without the fragmentation typically associated with short-read sequences. This capability is particularly crucial, as many bacterial genomes contain repetitive regions and substantial GC content, factors that have historically hampered full-length genome assemblies. The performance assessment conducted by researchers at BGI-Research aimed to fill these gaps presents raw data alongside processed outputs, thus ensuring transparency and reproducibility in scientific inquiry. This publication in the open-science journal GigaByte marks a significant milestone in the pursuit of assembling complete genomes.
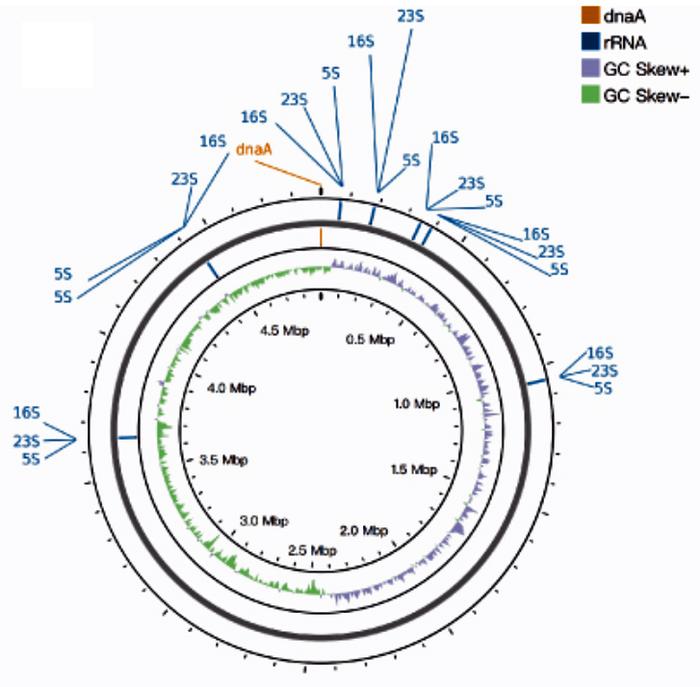
The benchmarking of CycloneSEQ™ was put to the test using Akkermansia muciniphila, a bacterial reference strain known for its importance in gut health. The evaluation yielded an impressive 12.07 Gbp of long-read data, presenting an average read length of 11.6 kbp. This hybrid approach, which combines the long-read capabilities of CycloneSEQ™ with the DNBSEQ™ short-read technology, achieved a complete assembly of the strain’s genome with an astonishingly low mismatch rate of less than 0.0001%. While traditional sequencing methods struggle with the complexities inherent in circular bacterial genomes, the CycloneSEQ™ platform has demonstrated that it can successfully navigate these challenges, producing reliable genomic assemblies with remarkable precision.
Moreover, this innovative sequencing technology was applied to a selection of ten bacterial strains isolated from human gut microbiota. Remarkably, researchers managed to close the genomes of all ten strains, including the additional circular structures of phages and plasmids that are frequently overlooked by conventional short-read sequencing. Through the employment of long-read only assemblies, complete genomes were constructed for eight out of the ten strains, marking a significant advancement over short-read-only methods, which failed to assemble any complete genomes from this selection.
Complex microbial communities also posed a testing ground for the CycloneSEQ™ platform. Through hybrid assembly techniques on a synthetic gut community consisting of 21 distinct microbial strains, researchers achieved five complete metagenome-assembled genomes (MAGs). This outsized performance contrast with both short-read and long-read methods underscores CycloneSEQ’s potential in unraveling the intricacies of microbial ecosystems, indicating its capability to handle diverse and complex samples that traditional approaches may falter upon.
The advantages of using long-read sequencing speak directly to its capacity for addressing the challenges of genome assembly. CycloneSEQ’s extended read lengths allow for the assembly of circular genomes, while DNBSEQ’s short-read technology supplements these assemblies by enhancing accuracy during the polishing phase. The ongoing research aims to explore additional non-synthetic samples, fine-tuning the integration between long and short reads in a bid to accelerate genome assembly while simultaneously ensuring quality.
An inherent challenge in microbiome studies has been the traditional methods’ limitations in resolving genomes from highly repetitive regions rich in GC content. CycloneSEQ™, with its direct sequencing approach that circumvents fragmentation issues, offers a solution to this long-standing problem. The study’s outcomes exemplify an evolving landscape in sequencing technology, one that effectively bridges the gaps left behind by the reliance on short-read methodologies.
As the field of genomics continues to advance, the implications of CycloneSEQ™ extend beyond mere sequencing capabilities. This platform not only aids in constructing accurate genomic representations but also adds layers of understanding to microbial functions and interactions within various environments. Its potential applications could reverberate through environmental monitoring, public health, and the development of novel therapeutic strategies targeting gut microbiota and other microbiomes.
The future landscape of sequencing technology is decidedly being reshaped by innovations such as CycloneSEQ™, and the scientific community stands at the precipice of a new era in which complete microbial genomes can be more routinely accessed and studied. The combination of extended read lengths, combined methodologies, and open access to data aligns with the increasingly collaborative nature of modern research, enhancing the capability for peer verification and scientific rigor.
As this technology matures and receives further validation through additional studies and comparative analyses, it is poised to pave the way for significant advancements in microbiology and genomics. Researchers are excited about the potential for CycloneSEQ™ to not only fill existing gaps in bacterial draft assemblies but also to enhance our understanding of microbial diversity, functionality, and evolutionary dynamics in a wide array of ecosystems.
In conclusion, CycloneSEQ™ represents a watershed moment in the field of genome sequencing, challenging the conventions established by earlier sequencing technologies. As researchers employ this advanced capability, the doors it opens for profound discoveries and advancements in microbial sciences are vast and promising.
Subject of Research: CycloneSEQ for Complete Bacterial Genomes
Article Title: Efficiently Constructing Complete Genomes with CycloneSEQ to Fill Gaps in Bacterial Draft Assemblies
News Publication Date: 25-Apr-2025
Web References: https://doi.org/10.1101/2024.09.05.611410
References: Hewei L, et al. Efficiently Constructing Complete Genomes with CycloneSEQ to Fill Gaps in Bacterial Draft Assemblies. GigaByte. 2025.
Image Credits: Hewei L, et al. Efficiently Constructing Complete Genomes with CycloneSEQ to Fill Gaps in Bacterial Draft Assemblies. GigaByte. 2025.
Keywords
Nanopore sequencing, Complete bacterial genomes, CycloneSEQ technology, Genome assembly, Hybrid sequencing methods, Microbial genomics, GigaScience, Long-read sequencing, DNBSEQ short-read technology, Gut microbiota.