Monoclonal antibodies are crucial therapeutics due to their high specificity and binding affinity for epitopes (the precise sites where an antibody attaches itself to an antigen, triggering an immune response). Since the therapeutic efficacy of antibodies is closely related to their target epitopes, epitope characterization is essential for understanding antibody functionality. However, the traditional methods used for studying epitopes are slow and labor-intensive. Therefore, there is a pressing need for more efficient approaches that can help map epitopes with precision and speed, ultimately accelerating the development of next-generation therapies.

Credit: Tokyo Tech
Monoclonal antibodies are crucial therapeutics due to their high specificity and binding affinity for epitopes (the precise sites where an antibody attaches itself to an antigen, triggering an immune response). Since the therapeutic efficacy of antibodies is closely related to their target epitopes, epitope characterization is essential for understanding antibody functionality. However, the traditional methods used for studying epitopes are slow and labor-intensive. Therefore, there is a pressing need for more efficient approaches that can help map epitopes with precision and speed, ultimately accelerating the development of next-generation therapies.
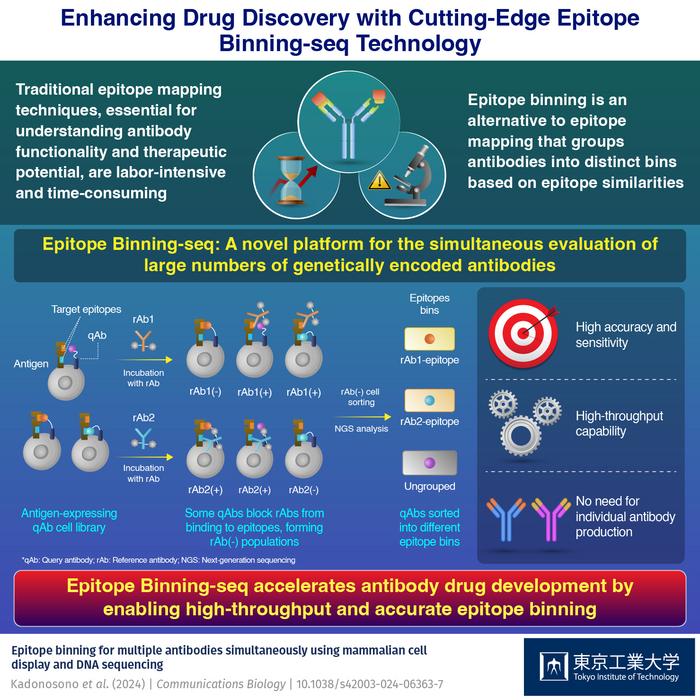
To bridge this gap, researchers from Tokyo Institute of Technology embarked on groundbreaking research, resulting in the development of a novel platform called Epitope Binning-seq. Their findings were published on 28 May, 2024 in Volume 7 of Communications Biology. Epitope Binning-seq, evaluates epitope similarity using genetically encoded query antibodies (qAbs) on antigen-expressing cells and next-generation sequencing (NGS), allowing simultaneous evaluation of multiple qAbs without individual purification.
In this study, query antibodies (qAbs) displayed on the surface of cells expressing HER2 were employed. By introducing fluorescently labeled reference antibodies (rAbs) and using flow cytometry analysis, researchers could distinguish between cells where the qAbs masked the epitopes and cells where they did not. Some qAbs effectively blocked the binding of rAb to the antigen, resulting in a population of cells with no rAb binding, termed the rAb-negative population. Conversely, other qAbs allowed the rAb to bind to the antigen, creating a population of cells with rAb binding, known as the rAb-positive population.
Giving further insights into their study, Associate Professor Tetsuya Kadonosono, the corresponding author of the study, says, “The differential binding behavior served as a basis for evaluating epitope similarity between different antibodies. We performed NGS analysis on the sorted rAb-negative populations to identify and group similar qAbs into epitope bins. This comprehensive approach enabled efficient analysis of a large number of antibodies and classification based on their epitope specificity.”
The results were quite promising. Epitope Binning-seq accurately classified antibodies into distinct epitope bins, providing valuable insights into their binding patterns. In experiments using model antibodies pertuzumab and trastuzumab, the method accurately identified and enriched specific qAbs, even detecting clones at very low initial abundances. The platform classified 14 qAbs into correct epitope bins, with high precision. These findings showcase Epitope Binning-seq’s potential to streamline early antibody drug development by evaluating millions of qAbs simultaneously, accelerating the identification of promising therapeutic candidates.
The benefits of this platform are significant. Epitope Binning-seq streamlines epitope comparison, enabling rapid identification of antibodies with similar binding patterns, potentially advancing targeted and effective antibody-based therapies. The method’s large-scale evaluation can transform antibody characterization, making it possible to assess millions of qAbs simultaneously.
“The development of Epitope Binning-seq represents a major advancement in antibody drug discovery. By providing a rapid, comprehensive, and cost-effective method for evaluating antibody epitope similarity, this novel platform overcomes the limitations of traditional epitope analysis techniques,” concludes Kadonosono.
The promising results of this study highlight the potential of this method to transform early antibody drug development and antibody affinity maturation, paving the way for more effective therapeutic strategies in the future.
###
About Tokyo Institute of Technology
Tokyo Tech stands at the forefront of research and higher education as the leading university for science and technology in Japan. Tokyo Tech researchers excel in fields ranging from materials science to biology, computer science, and physics. Founded in 1881, Tokyo Tech hosts over 10,000 undergraduate and graduate students per year, who develop into scientific leaders and some of the most sought-after engineers in industry. Embodying the Japanese philosophy of “monotsukuri,” meaning “technical ingenuity and innovation,” the Tokyo Tech community strives to contribute to society through high-impact research.
Journal
Communications Biology
Method of Research
Experimental study
Subject of Research
Not applicable
Article Title
Epitope binning for multiple antibodies simultaneously using mammalian cell display and DNA sequencing
Article Publication Date
28-May-2024