The human genome consists of around 3 billion base pairs and humans are all 99.6% identical in their genetic makeup. That small 0.4% accounts for any difference between one person and another. Specific combinations of mutations in those base pairs hold important clues about the causes of complex health issues, including heart disease and neurodegenerative diseases like schizophrenia.

Credit: Alexis Komor lab / UC San Diego
The human genome consists of around 3 billion base pairs and humans are all 99.6% identical in their genetic makeup. That small 0.4% accounts for any difference between one person and another. Specific combinations of mutations in those base pairs hold important clues about the causes of complex health issues, including heart disease and neurodegenerative diseases like schizophrenia.
Current methods to model or correct mutations in live cells are inefficient, especially when multiplexing — installing multiple point mutations simultaneously across the genome. Researchers from the University of California San Diego have developed new, efficient genome editing tools called multiplexed orthogonal base editors (MOBEs) to install multiple point mutations at once. Their work, led by Assistant Professor of Chemistry and Biochemistry Alexis Komor’s lab, appears in Nature Biotechnology.
Komor’s team was especially interested in comparing genomes that differ at a single letter change in the DNA. Those letters — C (cytosine), T (thymine), G (guanine), A (adenosine) – are known as bases. Where one person has a C base, another person might have a T base. These are single nucleotide variants (SNVs) or single point mutations, a person might have 4-5 million variants. Some variants are harmless; some are harmful; and often it is a combination of variants that confers disease.
One issue with using the genome in disease modeling is the sheer number of possible variations. If scientists were trying to determine which genetic mutations were responsible for heart disease, they could decode the genomes of a cohort that all had heart disease but the number of variations between any two people makes it very hard to determine which combination of variations causes the disease.
“There is a problem interpreting genetic variants. In fact, most variants that are identified are unclassified clinically, so we don’t even know if they’re pathogenic or benign,” stated Quinn T. Cowan, a recent Ph.D. graduate from the university’s Department of Chemistry and Biochemistry and first author on the paper. “Our goal was to make a tool that can be used in disease modeling by installing multiple variants in a controlled laboratory setting where they can be studied further.”
An evolution in gene-editing
To understand why MOBEs were created, we have to understand the limitations of the traditional gene-editing tool CRISPR-Cas9. CRISPR-Cas9 uses a guide RNA, which acts like a GPS signal that goes straight to the genomic location you want to edit. Cas9 is the DNA-binding enzyme that cuts both strands of the DNA, making a complete break.
Although relatively straightforward, double-stranded breaks can be toxic to cells. This kind of gene-editing can also lead to indels — random insertions and deletions — where the cell is not able to perfectly repair itself. Editing multiple genes in CRISPR-Cas9 multiples the risks.
Instead of CRISPR, Komor’s lab uses a base-editing technique she developed, which makes a chemical change to the DNA, although only one type of edit (C to T or A to G, for example) can be made at a time. So rather than scissors that cut out an entire section at once, base-editing erases and replaces one letter at a time. It is slower, but more efficient and less harmful to cells.
Simultaneously applying two or more base editors (changing a C to T at one location, and an A to G at another location in the genome), allows for better modeling of polygenic diseases — those occurring due to more than one genetic variant. However, a technology didn’t exist that could do this efficiently without guide RNA “crosstalk,” which happens when base editors make unwanted changes.
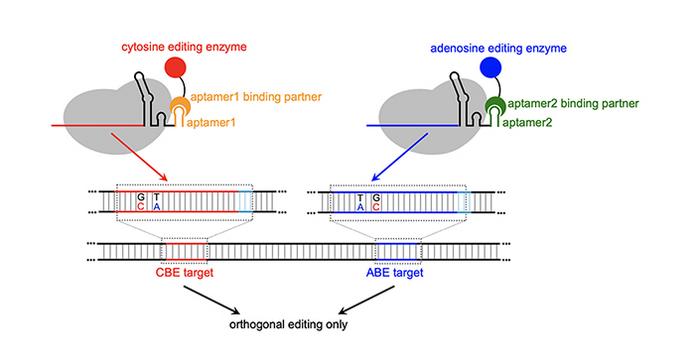
Cowan’s MOBEs use RNA structures called aptamers — small RNA loops that bind to specific proteins — to recruit base-modifying enzymes to specific genomic locations enabling simultaneous editing of multiple sites with high efficiency and a lower incidence of crosstalk.
This system is novel and is the first time someone used aptamers to recruit ABEs (adenosine base editors) in combination with CBEs (cytosine base editors) in an orthogonal pattern to make the MOBEs.
The differences are stark: when CBE and ABE are given together not using MOBE, crosstalk occurs up to 30% of the time. With MOBE, crosstalk is less than 5%, while achieving 30% conversion efficiency of the desired base changes.
The study was a proof of principle to test the feasibility of the MOBE system, which has been granted a provisional patent. To test them even further, the team conducted several case studies with real diseases, including Kallmann syndrome, a rare hormonal disorder. Their experiments revealed that MOBE systems could be used to efficiently edit relevant cell lines of certain polygenic diseases.
“We’re in the process of putting the plasmids up on AddGene so anyone can freely access them. Our hope is that other researchers will use the MOBEs to model genetic diseases, learn how they manifest and then hopefully create effective therapies,” stated Cowan.
This research was funded in part by the National Institutes of Health (1R35GM138317, T32 GM008326, and T32 GM112584) and the Research Corporation for Science Advancement (28385).
Full list of authors: Quinn T. Cowan, Sifeng Gu, Wanjun Gu, Brodie L. Ranzau, Tatum S. Simonson, and Alexis C. Komor (all UC San Diego).
Journal
Nature Biotechnology
Method of Research
Experimental study
Article Title
Development of multiplexed orthogonal base editor (MOBE) systems
Article Publication Date
21-May-2024