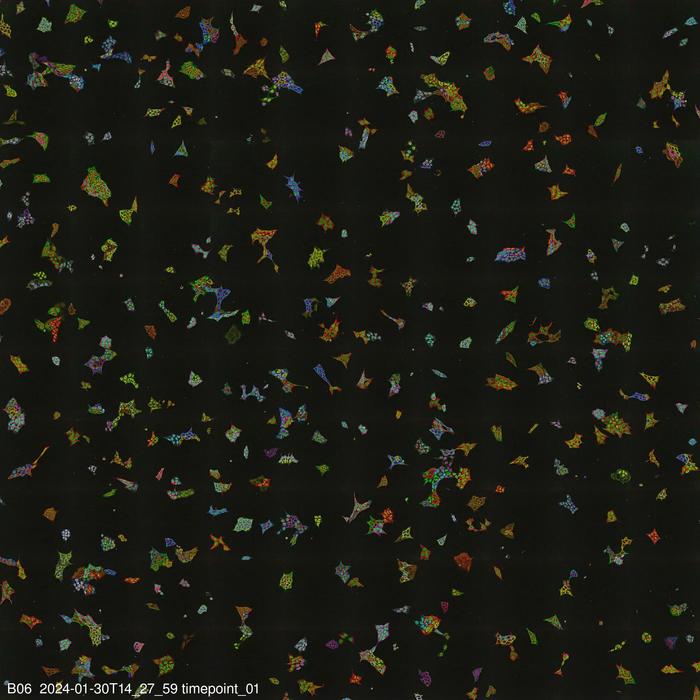Observing proteins precisely within cells is extremely important for many branches of research but has been a significant technical challenge – especially in living cells, as the required fluorescent labelling had to be individually attached to each protein. The research group led by Stefan Kubicek at CeMM has now overcome this hurdle: With a method called “vpCells,” it is possible to label many proteins simultaneously, using five different fluorescent colours. This automated high-throughput approach, aided by AI-assisted image recognition, opens up entirely new applications in various disciplines, from fundamental cell biology to drug discovery. The study has been published in the journal Nature Cell Biology (DOI: 10.1038/s41556-024-01407-w), and the related images are available at vpcells.cemm.at.

Credit: (c) Andreas Reicher/Jiri Reinis
Observing proteins precisely within cells is extremely important for many branches of research but has been a significant technical challenge – especially in living cells, as the required fluorescent labelling had to be individually attached to each protein. The research group led by Stefan Kubicek at CeMM has now overcome this hurdle: With a method called “vpCells,” it is possible to label many proteins simultaneously, using five different fluorescent colours. This automated high-throughput approach, aided by AI-assisted image recognition, opens up entirely new applications in various disciplines, from fundamental cell biology to drug discovery. The study has been published in the journal Nature Cell Biology (DOI: 10.1038/s41556-024-01407-w), and the related images are available at vpcells.cemm.at.
Without proteins, life as we know it would be inconceivable. They provide the structural framework for cells, act as enzymes to control metabolism, and enable cells to communicate with their environment as membrane receptors, transporters, or signalling molecules. All of these functions can only be fulfilled if the proteins are located in the right place within the cell. Often, even the properties of a protein change when it changes its location – control over its localization in the cell therefore also means control over its function.
To understand and explore the function of proteins, it is essential to precisely determine and track their location within the cell. Proteins often shuttle dynamically between different organelles and compartments of the cell. To visualize them under the microscope, they are often linked to a fluorescent, brightly shining protein component. However, this method has faced technical difficulties: Typically, the fluorescent component could only be attached to one protein at a time, and to label multiple proteins, cells usually had to be killed and fixed.
The new method presented by Stefan Kubicek’s group, called “visual proteomics Cells” (abbreviated vpCells), allows proteins to be fluorescently labelled in a way that preserves their endogenous regulatory mechanisms. Instead of labelling one protein at a time, vpCells can fuse many proteins simultaneously with a fluorescent tag in a so-called multiplex approach. A precursor of this method was already described by Kubicek’s team in 2020 for studying metabolic enzymes (Reicher et al. Genome Res. 2020, DOI: 10.1101/gr.261503.120). Now it has been expanded and improved in three ways:
Firstly, vpCells can label all theoretically possible proteins using the CRISPR/Cas9 gene-editing tool to genetically attach fluorescent proteins to the proteins under investigation. Kubicek’s group has created a genome-wide “library” for this purpose, enabling the fluorescent marking and systematic functional exploration of all possible human proteins.
Secondly, vpCells use not only one fluorescent colour but a total of five complementary colours. In each cell, two different proteins to be tracked are marked. Additionally, another colour marking is used to better distinguish individual clones. And two further colours mark the cell nucleus and membrane to delineate individual cells better.
Thirdly, this colour scheme enables not only to generate visually appealing images, but also to optically recognize and discriminate the different proteins . Normally, this requires complex DNA sequencing after imaging to determine which protein is labelled. The vpCells approach, on the other hand, enables training an AI-assisted image recognition system to recognize which protein is marked in which cell based solely on fluorescence microscopy images.
The method has already demonstrated its utility in two applications: On the one hand, more than 4,500 cell lines were generated as reporters for more than 1,100 proteins. These cell lines were used to train the AI models and to describe localization of the proteins in their basal state. All images of the individual labelled proteins are available on the publicly accessible web database vpcells.cemm.at.
On the other hand, the living reporter cells were used for a specific research question: Kubicek’s team examined the effect of more than 1000 small-molecule substances on 61 proteins relevant to cancer cells. The researchers found that 44 of the tested substances altered the amount or localization of individual proteins within 6 hours. One of the substances turned out to be an inhibitor of protein transport from the cell nucleus, which has a similar effect to a clinically approved drug for multiple myeloma, a cancer of the blood-forming system.
“These results provide a first glimpse into the versatility of the vpCells method,” says Kubicek. “We expect many more future applications , from fundamental cell biology to applied drug discovery.”
***
The Study “Pooled multicolor tagging for visualizing subcellular protein dynamics” was published in Nature Cell Biology on April 19, 2024 . DOI: 10.1038/s41556-024-01407-w
Authors: Andreas Reicher, Jiří Reiniš, Maria Ciobanu, Pavel Růžička, Monika Malik, Marton Siklos, Viktoriia Kartysh, Tatjana Tomek, Anna Koren, André F. Rendeiro, Stefan Kubicek
Funding: Andreas Reicher was supported by a Boehringer Ingelheim Fonds PhD fellowship, André Rendeiro by Angelini Ventures S.p.A. Rome, Italy. Research in the Kubicek lab has been supported by the Austrian Academy of Sciences, the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program, the Austrian Science Fund (FWF), and the Vienna Science and Technology Fund (WWTF).
Stefan Kubicek joined CeMM in 2010. He obtained an MSc in Synthetic Organic Chemistry from the Vienna University of Technology after writing a diploma thesis at ETH Zurich. For his PhD in Thomas Jenuwein’s lab at the IMP in Vienna, he changed fields to molecular biology and developed the first selective histone methyl transferase inhibitors. He then performed postdoctoral research, working on chemical biology with Stuart Schreiber at the Broad Institute of Harvard and MIT. At CeMM, Stefan Kubicek headed the Christian Doppler Laboratory for Chemical Epigenetics and Antiinfectives, a public-private partnership between CeMM, Boehringer Ingelheim, and Haplogen, and is now the head of the CeMM Molecular Discovery Platform. The Kubicek lab is working on the role of chromatin in the definition of cell types and cell states, in particular chromatin-modifying enzymes as synthetic lethal targets in cancer and chemical transdifferentiation to insulin-producing beta cells. In an ERC-funded project, the laboratory studies metabolic enzymes in the cell’s nucleus to test the hypothesis that small molecule metabolites shape chromatin structure and thus control gene expression and cell identity.
The CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences is an international, independent and interdisciplinary research institution for molecular medicine under the scientific direction of Giulio Superti-Furga. CeMM is oriented towards medical needs and integrates basic research and clinical expertise to develop innovative diagnostic and therapeutic approaches for precision medicine. Research focuses on cancer, inflammation, metabolic and immune disorders, and rare diseases. The Institute’s research building is located on the campus of the Medical University and the Vienna General Hospital.
For further information please contact:
Stefan Bernhardt
PR & Communications Manager
CeMM
Research Center for Molecular Medicine
of the Austrian Academy of Sciences
Lazarettgasse 14, AKH BT 25.3
1090 Vienna, Austria
Phone +43-1/40160-70 056
Fax +43-1/40160-970 000
sbernhardt@cemm.at
Journal
Nature Cell Biology
Method of Research
Imaging analysis
Subject of Research
Cells
Article Title
Pooled multicolor tagging for visualizing subcellular protein dynamics
Article Publication Date
19-Apr-2024