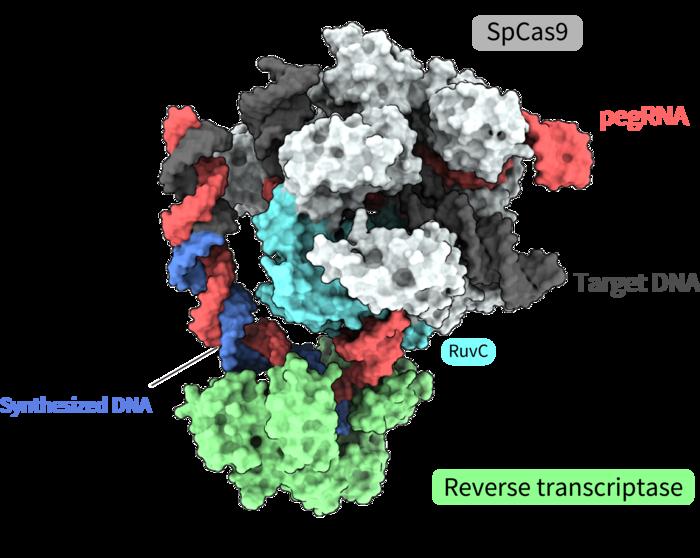
Joint research led by Yutaro Shuto, Ryoya Nakagawa, and Osamu Nureki of the University of Tokyo determined the spatial structure of various processes of a novel gene-editing tool called “prime editor.” Functional analysis based on these structures also revealed how a “prime editor” could achieve reverse transcription, synthesizing DNA from RNA, without “cutting” both strands of the double helix. Clarifying these molecular mechanisms contributes greatly to designing gene-editing tools accurate enough for gene therapy treatments. The findings were published in the journal Nature.

Credit: Shuto et al 2024
Joint research led by Yutaro Shuto, Ryoya Nakagawa, and Osamu Nureki of the University of Tokyo determined the spatial structure of various processes of a novel gene-editing tool called “prime editor.” Functional analysis based on these structures also revealed how a “prime editor” could achieve reverse transcription, synthesizing DNA from RNA, without “cutting” both strands of the double helix. Clarifying these molecular mechanisms contributes greatly to designing gene-editing tools accurate enough for gene therapy treatments. The findings were published in the journal Nature.
The 2020 Nobel Prize in Chemistry was awarded to Jennifer Doudna and Emmanuelle Charpentier for developing a groundbreaking yet simple way to edit DNA, the “blueprint” of living organisms. While their discovery opened new avenues for research, the accuracy of the method and safety concerns about “cutting” both strands of DNA limited its use for gene therapy treatments. As such, research has been underway to develop tools that do not have these drawbacks.
The prime editing system is one such tool, a molecule complex consisting of two components. One component is the prime editor, which combines a SpCas9 protein, used in the first CRISPR-Cas gene editing technology, and a reverse transcriptase, an enzyme that transcribes RNA into DNA. The second component is the prime editing guide RNA (pegRNA), a modified guide RNA that identifies the target sequence within the DNA and encodes the desired edit. In this complex, the prime editor works like a “word processor,” accurately replacing genomic information. The tool has already been successfully implemented in living cells of organisms such as plants, zebrafish, and mice. However, precisely how this molecule complex executes each step of the editing process has not been clear, mostly due to a lack of information on its spatial structure.
“We became curious about how the unnatural combination of proteins Cas9 and reverse transcriptase work together,” says Shuto, the first author of the paper.
The research team used cryogenic electron microscopy, an imaging technique that makes observations possible at a near-atomic scale. The method required samples to be in glassy ice to protect them from the potential damage by the electron beams, posing some additional challenges.
“We found the prime editor complex to be unstable under experimental conditions,” explains Shuto. “So, it was very challenging to optimize the conditions for the complex to stay stable. For a long time, we could only determine the structure of Cas9.”
Finally overcoming the challenges, the researchers succeeded in determining the three-dimensional structure of the prime editor complex in multiple states during reverse transcription on the target DNA. The structures revealed that the reverse transcriptase bound to the RNA–DNA complex that formed along the “part” of the Cas9 protein associated with DNA cleavage, the splitting of a single strand of the double helix. While performing the reverse transcription, the reverse transcriptase maintained its position relative to the Cas9 protein. The structural and biochemical analyses also indicated that the reverse transcriptase could lead to additional, undesired insertions.
These findings have opened new avenues for both basic and applied research. So, Shuto lays out the next steps.
“Our structure determination strategy in this study can also be applied to prime editors composed of a different Cas9 protein and reverse transcriptase. We want to utilize the newly obtained structural information to lead to the development of improved prime editors.”
Journal
Nature
Method of Research
Experimental study
Subject of Research
Not applicable
Article Title
Structural basis for pegRNA-guided reverse transcription by the prime editor
Article Publication Date
29-May-2024