In the rapidly evolving field of cancer genomics, long non-coding RNAs (lncRNAs) have become a focal point of research due to their profound regulatory roles in gene expression and tumor biology. A groundbreaking study recently published in the open-access journal Gene Expression has shed new light on the differential expression of lncRNAs within peripheral blood mononuclear cells (PBMCs) of women diagnosed with luminal A breast cancer. This subtype, known for its hormone receptor positivity and relatively favorable prognosis, nonetheless requires improved diagnostic and prognostic biomarkers for early detection and therapeutic intervention. By harnessing advanced microarray technology and rigorous bioinformatic analyses, researchers have identified specific lncRNAs with significant potential as minimally invasive biomarkers, signaling a promising leap forward in breast cancer diagnostics.
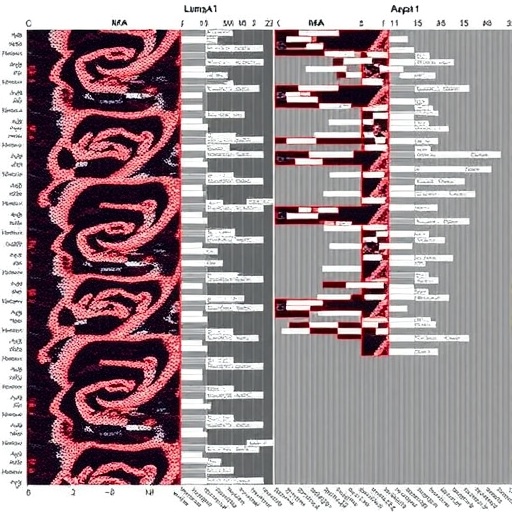
The study employed a one-color microarray platform, utilizing SurePrint G3 Human Unrestricted 8×60K arrays paired with Agilent’s SureScan Microarray Scanner, facilitating extensive transcriptome-wide profiling of PBMCs. The selection of PBMCs as a source of genetic material was strategic, capitalizing on their accessibility through peripheral blood draws and their reflective capacity of systemic pathological states. The cohort consisted of sixteen subjects, evenly divided between patients with luminal A breast cancer and matched healthy controls, ensuring a controlled comparative framework. Subsequently, the team applied the robust “limma” package alongside the versatile “tidyverse” suite in the R environment to identify differentially expressed lncRNAs with statistical stringency, controlling for false discovery rates to mitigate type I errors.
Results highlighted significant dysregulation of several lncRNA classes, notably long intergenic non-coding RNAs (LINC), LOC genes, and antisense transcripts. Of particular interest was LINC00974, which exhibited a marked increase in expression in cancer patients compared to controls, with a log fold change exceeding 1.5 and an FDR-adjusted p-value of 0.03. This rigorously validated differential expression underscores LINC00974’s potential as a sensitive and specific biomarker for early-stage breast cancer detection. The biological significance of LINC00974 is supported by previous literature elucidating its role in oncogenic pathways, primarily through mechanisms involving microRNA sponging—a process that modulates availability of miRNAs, consequently regulating downstream gene expression patterns pivotal in cell proliferation, migration, and tumor metastasis.
Fascinatingly, the functional enrichment analysis revealed that differentially expressed lncRNAs cluster into gene networks linked to oncogenesis and tumor progression. The integration of findings from the LncRNADisease 2.0 database further confirmed associations between these lncRNAs and diverse oncological disorders, suggesting a shared molecular regulatory framework underpinning multiple cancer types. This cross-cancer relevance amplifies the translational potential of targeting such lncRNAs, not only as diagnostic markers but also as therapeutic candidates, offering a novel axis for precision medicine approaches.
The discovery that lncRNA alterations are detectable in PBMCs, peripheral blood cells, is particularly noteworthy. This finding supports the concept that systemic blood components mirror tumor-derived molecular signatures, circumventing the need for invasive tissue biopsies. It opens avenues for blood-based liquid biopsy tests, which could revolutionize breast cancer screening by providing a simple, non-invasive, and repeatable method for early diagnosis and monitoring. Considering the aggressive nature of breast cancer metastasis and the importance of early intervention for favorable outcomes, such biomarker development is urgently needed.
Importantly, LINC00974’s involvement in chromatin remodeling and RNA stabilization provides mechanistic insights into how non-coding RNAs orchestrate complex regulatory networks within the tumor microenvironment and circulating immune cells alike. These processes influence the epigenetic landscape and post-transcriptional control of gene expression, directly impacting tumor cell behavior and immune responses. Understanding these pathways could unravel new targets for pharmaceutical modulation and shed light on resistance mechanisms to conventional therapies.
The study’s limitations, acknowledged by the authors, include the relatively small sample size, which, while sufficient for exploratory analysis, necessitates validation in larger cohorts to corroborate these findings and establish clinical utility. Future work will focus on functional assays to confirm the biological roles of these candidate lncRNAs and refine their specificity and sensitivity profiles. Techniques such as quantitative PCR will be employed to validate expression levels independently, ensuring robustness of the biomarker candidates.
A compelling direction for upcoming research is the longitudinal monitoring of lncRNA expression changes through treatment and disease progression. Such dynamic profiling could enable personalized therapeutic adjustments and provide prognostic information, potentially identifying patients at higher risk of relapse or metastasis. It also aligns with emerging trends in oncology toward integrating molecular diagnostics with patient management, fostering a move toward precision health.
The implications of this research extend beyond breast cancer, as the molecular principles governing lncRNA function appear conserved across multiple cancer types. This lends weight to the hypothesis that lncRNAs contribute to the hallmarks of cancer and represent a largely untapped reservoir of molecular targets. The intersection of non-coding RNA biology with immunology, as illustrated by PBMC analyses, may uncover novel avenues to modulate immune surveillance and tumor-immune interactions.
Moreover, the methodology showcased in this study exemplifies the power of combining high-throughput technologies with sophisticated computational tools to unveil subtle yet clinically meaningful molecular alterations. The study integrates bioinformatics pipelines adept at multiple testing correction and functional enrichment, highlighting best practices in omics research for reliable biomarker discovery.
In summary, this pioneering investigation elucidates the altered landscape of long non-coding RNAs in peripheral blood mononuclear cells of luminal A breast cancer patients, underscoring LINC00974 as a frontrunner biomarker candidate. Its detectability in blood and involvement in oncogenic pathways position it as a potential game-changer in early cancer detection and targeted therapy development. As subsequent studies expand upon these findings, the vision of minimally invasive, lncRNA-based diagnostic assays for breast cancer edges closer to reality, promising to enhance patient outcomes through timely intervention and personalized care.
Subject of Research: Long non-coding RNAs in peripheral blood mononuclear cells associated with luminal A breast cancer
Article Title: Non-coding RNAs in Peripheral Blood Mononuclear Cells in Luminal A Breast Cancer
News Publication Date: 13-Aug-2025
Web References:
- Journal: Gene Expression
- DOI: 10.14218/GE.2025.00021
Keywords: Long noncoding RNA, Breast cancer, Luminal A, Peripheral blood mononuclear cells, LINC00974, Biomarkers, Microarray analysis, Oncogenic pathways, miRNA sponging, Gene expression regulation