Understanding the relationship between mtDNA and animal growth could provide valuable information for selective breeding in aquaculture. However, the complex interactions between genetics and environmental factors often hinders progress in this field. To that end, a recent study published in the KeAi hournal Reproduction and Breeding investigated the cross-sectional diameter of skeletal muscle fibers in allotriploid fish with different growth traits.

Credit: Zhang, H., et al.
Understanding the relationship between mtDNA and animal growth could provide valuable information for selective breeding in aquaculture. However, the complex interactions between genetics and environmental factors often hinders progress in this field. To that end, a recent study published in the KeAi hournal Reproduction and Breeding investigated the cross-sectional diameter of skeletal muscle fibers in allotriploid fish with different growth traits.
The researchers selected red crucian carp, common carp and two allotriploid fish for the study. The allotriploid fish exhibit distinct growth rates and ploidy levels compared to the inbred parental species (red crucian carp and common carp). Deciphering the genetic mechanisms underlying these growth differences holds significant potential for fish breeding.
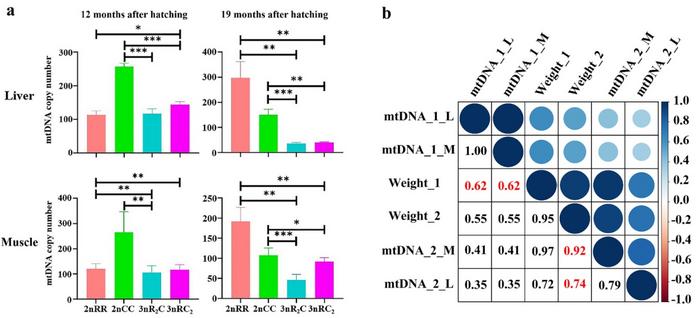
“Distant hybridization rapidly generates diverse genotypes and phenotypes, offering a rich resource for studying the role of genetic regulation in shaping phenotypes,” explains co-corresponding author Li Ren, a researcher at the State Key Laboratory of Developmental Biology of Freshwater Fish at Hunan Normal University. “Analysis of muscle fiber diameter and mtDNA copy number revealed a positive correlation between individual size and muscle fiber diameter across all fish types. Additionally, a strong correlation emerged between muscle mtDNA copy number and body weight.”
Notably, the researchers observed a decrease in muscle mtDNA copy number within the allotriploid fish during winter. “This decrease likely corresponds with reduced individual activity and energy expenditure aimed at maintaining body weight, potentially contributing to the observed growth heterosis of allotriploid fish,” adds Ren.
Furthermore, the researchers analyzed the expression levels of three nuclear-regulated mitochondrial genes (tfam, tfb1m, and tfb2m). Interestingly, they found imbalanced allelic expression of tfam and tfb1m within the allotriploid fish. Ren highlights the complex relationship between mtDNA, “The copy number and transcriptional efficiency of animal mtDNA are intimately linked not only to the characteristics of the mtDNA genome itself but also to the expression of specific nuclear genes. However, at present, a direct link between these three nuclear-encoded mitochondrial genes and mtDNA copy number variation remains elusive.”
###
Contact the author: Li Ren, State Key Laboratory of Developmental Biology of Freshwater Fish, School of Life Sciences, Hunan Normal University, Changsha, 410081, Hunan, China. renli_333@163.com.
The publisher KeAi was established by Elsevier and China Science Publishing & Media Ltd to unfold quality research globally. In 2013, our focus shifted to open access publishing. We now proudly publish more than 100 world-class, open access, English language journals, spanning all scientific disciplines. Many of these are titles we publish in partnership with prestigious societies and academic institutions, such as the National Natural Science Foundation of China (NSFC).
Journal
Reproduction and Breeding
Method of Research
Experimental study
Subject of Research
Animals
Article Title
mtDNA copy number contributes to growth diversity in allopolyploid fish
COI Statement
The authors declare that they have no competing interests.