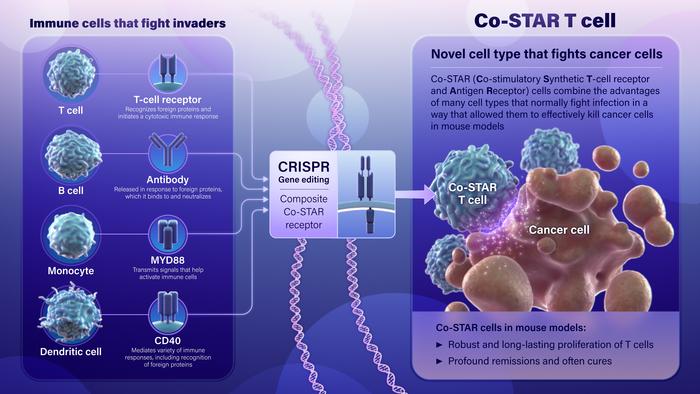
Using genetic engineering techniques, investigators at the Johns Hopkins Kimmel Cancer Center and its Ludwig Center, the Lustgarten Laboratory and Bloomberg~Kimmel Institute for Cancer Immunotherapy have designed a novel type of cell to recognize and fight cancer.

Credit: Elizabeth Cook
Using genetic engineering techniques, investigators at the Johns Hopkins Kimmel Cancer Center and its Ludwig Center, the Lustgarten Laboratory and Bloomberg~Kimmel Institute for Cancer Immunotherapy have designed a novel type of cell to recognize and fight cancer.
To produce the cells, called Co-STAR (Co-stimulatory Synthetic T-cell receptor and Antigen Receptor) cells, the researchers combined genetic components of four types of cells that the body normally uses to defend against invaders to make a powerful new cell type: T-cell receptors (TCRs) from T cells, antibodies from B cells, MyD88 from white blood cells called monocytes, and CD40 from dendritic and other cells. The TCR and antibody components served as an “invader detecting device,” recognizing cancer cells as foreign, and the “alarm” triggered by this hybrid detector was boosted by the MyD88 and C40 components.
In laboratory studies, Co-STARs led to a sustained anti-tumor response against human cancer cells growing in test tubes and in mice. A description of the work was published July 10 in Science Translational Medicine.
T cell-based therapies are among the most promising approaches to treat advanced cancer and are the subject of intense research, explains lead study author Brian Mog, M.D., Ph.D., an internal medicine resident at Brigham and Women’s Hospital in Boston. He was a medical and graduate student at the Johns Hopkins University School of Medicine when the research was conducted.
However, TCR and CAR (chimeric antigen receptor, usually using antibody as the detector), which are aimed at stimulating an immune response by activating T cells, each has limits. The combination of the two can overcome these limitations.
“We needed to make a new type of cell, because we were trying to target specific antigens called peptide-HLA (human leukocyte antigen) antigens, which are peptide fragments from mutant proteins inside the cancer cell that are displayed on the cell surface by peptide-holding proteins called HLAs,” Mog explains. Their specific target was a peptide containing the R175H mutation of p53 (the 175th amino acid of p53 is mutated from arginine to histidine), displayed on the HLA-A2 allele (gene variation). This is the most common mutation in the tumor suppressor protein p53, which is in turn the most commonly mutated gene in human cancers.
However, these antigens are present at very low numbers (just one to 10) on a cancer cell, and the classic CAR format would not be able to react to such a small amount. “Our goal was to combine some of the advantages of the CAR format with those of the natural T cell receptor on T cells, supplemented with additional signaling boosters, so that they could fight cancers more effectively,” Mog says.
The team went through multiple rounds of engineering to come up with the final design, testing their receptors in model cancer cell lines in test tubes and then in mouse models of cancer. The final Co-STAR T cells were able to continuously kill human cancer cells in test tubes. When tested in mouse models of cancer, Co-STARs induced a robust, long-lasting proliferation of T cells that were able to induce profound remissions, and often cure, human cancer cells growing in mice. By contrast, more conventional T cells or CAR T cells were not able to eradicate the cancer cells in vitro and only brought about temporary tumor control in mice, with the cancers re-emerging days later.
“Brian’s results demonstrated that Co-STAR T cells combine the advantages of many features of immune cells that normally fight infection in a way that allowed them to effectively kill cancer cells in mouse models,” says co-senior investigator Bert Vogelstein, M.D., Clayton Professor of Oncology, Howard Hughes Medical Institute investigator and co-director of the Ludwig Center. “Co-STARs address some, but certainly not all, challenges confronting T cell-based therapeutics but are certainly worthy of continued investigation.”
“I was, honestly, incredibly surprised that the Co-STARs worked so well in mice, given that I had generated so many different types of T cells over four years that could only slow the growth of cancers in mice” adds Mog. “Witnessing those cures was a very exciting moment.”
Study co-authors were Nikita Marcou, Sarah DiNapoli, Alexander Pearlman, Tushar Nichakawade, Michael Hwang, Jacqueline Douglass, Emily Han-Chung Hsiue, Stephanie Glavaris, Katharine Wright, Maximilian Konig, Suman Paul, Nicolas Wyhs, Jiaxin Ge, Michelle Miller, P. Aitana Azurmendi, Evangeline Watson, Drew Pardoll, Sandra Gabelli, Chetan Bettegowda, Nickolas Papadopoulos, Kenneth Kinzler and Shibin Zhou of Johns Hopkins.
The work was supported by the Virginia and D.K. Ludwig Fund for Cancer Research, the Lustgarten Foundation, the Commonwealth Fund, Bloomberg Philanthropies and the Bloomberg~Kimmel Institute for Cancer Immunotherapy, the National Institutes of Health (NH) Cancer Center Support Grant P30 CA006973, NIH grants (T32 GM136577, T32 AR048522, 1R21 AI176764, and K08CA270403), the National Institute of General Medical Sciences (grant T32GM148383), the National Cancer Institute (grants T32CA153952 and R37CA230400) and awards from the Leukemia & Lymphoma Society Translational Research Program, the American Society of Hematology, Swim Across America translational cancer research, the Jerome Greene Foundation, the Cupid Foundation, the Stephen and Renee Bisciotti Foundation, the Harrington Scholar-Innovator grant and the Rheumatology Research Foundation.
The Johns Hopkins University has filed patent applications related to technologies described in this paper on which Hsiue, Wright, Douglass, Mog, Hwang, Pearlman, Papadopoulos, Kinzler, Vogelstein, Gabelli, Pardoll and Zhou are listed as inventors. Vogelstein, Kinzler and Papadopoulos are founders of (and Kinzler and Papadopoulos are consultants to) Thrive Earlier Detection, an Exact Sciences company. Vogelstein, Kinzler, Papadopoulos and Zhou hold equity in Exact Sciences and are founders of or consultants to and own equity in Clasp Therapeutics, NeoPhore, and Personal Genome Diagnostics. Vogelstein, Kinzler and Papadopoulos are founders of or consultants to and own equity in Haystack Oncology and Cage Pharma. Papadopoulos is a consultant to Vidium. Vogelstein is a consultant to and holds equity in Catalio Capital Management. Zhou has a research agreement with BioMed Valley Discoveries. Bettegowda is a consultant to DePuy Synthes, Bionaut Labs, Haystack Oncology, Galectin Therapeutics, and Privo Technologies, and is a co-founder of OrisDX and Belay Diagnostics. Gabelli is a founder and holds equity in AMS LLC. Konig received consulting fees from Argenx, Atara Biotherapeutics, Revel Pharmaceuticals, Sana Biotechnology and Sanofi. Douglass previously consulted for Hemogenyx Pharmaceuticals. Paul is a consultant for Merck, owns equity in Gilead and received payment from IQVIA and Curio Science. Pardoll reports grant and patent royalties through Johns Hopkins from BMS, a grant from Compugen, stock from Trieza Therapeutics and Dracen Pharmaceuticals, and founder equity from Potenza. He is a consultant for Aduro Biotech, Amgen, AstraZeneca (MedImmune/Amplimmune), Bayer, DNAtrix, Dynavax Technologies, Ervaxx, FLX Bio, Rock Springs Capital, Janssen, Merck, Tizona and Immunomic Therapeutics. Pardoll is also on the scientific advisory boards of Five Prime Therapeutics, Camden Nexus II and WindMIL and the board of directors for Dracen Pharmaceuticals. Wright and Gabelli are current or former employees of Merck Sharp & Dohme and may own stock or stock options. The companies named above, as well as others, have licensed previously described technologies related to the work described in this paper from The Johns Hopkins University. Vogelstein, Kinzler and Papadopoulos are inventors of some of these technologies. Licenses for these technologies are or will be associated with equity or royalty payments to the inventors as well as to The Johns Hopkins University. Patent applications for the work described in this paper may be filed by The Johns Hopkins University. The terms of all of these arrangements are being managed by The Johns Hopkins University in accordance with its conflict-of-interest policies.
Journal
Science Translational Medicine