In the rapidly evolving landscape of neurodegenerative disease research, a groundbreaking study published in npj Parkinson’s Disease details the development of cutting-edge computational tools designed to revolutionize histopathological analysis of synucleinopathies in mouse models. Employing convolutional neural networks (CNNs), a sophisticated form of deep learning technology, this novel approach enables fully automated, brain-wide examination of pathological changes, marking a transformative advancement in the study of Parkinson’s disease and related disorders.
At the core of this innovation lies the utilization of CNNs, which have been trained extensively to recognize specific histopathological hallmarks associated with synuclein-related neurodegeneration. Traditional pathological analysis in this realm has been labor-intensive, highly subjective, and prone to variability, hindering large-scale and reproducible results. By automating this process, the study surmounts prevalent limitations through unbiased, high-throughput analysis with unprecedented spatial resolution throughout the brain.
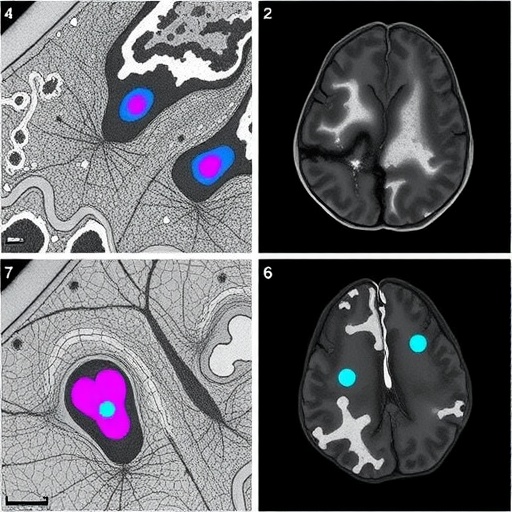
The methodology implemented by Barber-Janer and colleagues integrates high-resolution histological imaging with deep learning architectures tailored to parse complex morphological patterns. The CNN was optimized to detect alpha-synuclein aggregates, a defining pathological proteinopathy in Parkinson’s disease. This protein misfolding and aggregation cascade is a critical feature underpinning synucleinopathies, making its accurate identification essential for both diagnostic and therapeutic research.
Importantly, the study’s neural networks were trained on meticulously annotated datasets derived from well-characterized mouse models genetically engineered to express synucleinopathy phenotypes. This training regimen enhanced the algorithm’s ability to generalize across diverse pathological manifestations, ensuring robust performance despite biological variability. The researchers benchmarked the CNN outputs against expert neuropathologist assessments, demonstrating a high concordance rate and thus validating the model’s practical utility.
One of the most remarkable achievements of this work is the ability to perform brain-wide mapping of pathological burden. By automating this process, the researchers could quantify and visualize spatial distribution patterns of alpha-synuclein deposits throughout different brain regions in three dimensions. Such comprehensive mapping facilitates deeper insights into disease progression, neuroanatomic vulnerability, and potential pathways for therapeutic intervention.
Beyond detection, the CNN’s analytical capacity extends to distinguishing between diverse morphological phenotypes of alpha-synuclein aggregates, ranging from small punctate inclusions to larger, more complex Lewy body-like formations. This capability introduces a new level of granularity to neuropathological studies, allowing researchers to investigate correlations between aggregate morphology and disease severity or stage.
The implications of this automation transcend translational research alone. The platform promises to accelerate preclinical therapeutic screening by providing rapid, objective readouts of disease-modifying effects across various treatment paradigms. This can significantly streamline drug development pipelines, ultimately hastening clinical translation efforts for Parkinson’s disease and related neurodegenerative disorders.
Furthermore, the open-source nature of the developed CNN framework inspires collaborative enhancement by the scientific community. Researchers worldwide can adapt and refine the model for application in other proteinopathies or experimental conditions. The scalability of this approach underscores its potential as a universal tool for histopathological analysis in neurodegeneration research.
Technical innovations underpinning the study include the deployment of advanced image preprocessing pipelines, facilitating artifact correction and normalization to optimize input quality for deep learning inference. The multi-scale architecture of the CNN, incorporating layers adept at capturing both micro and macro-anatomical features, represents a sophisticated integration of computational design tailored to biological complexity.
Statistical validation involved rigorous cross-validation techniques and performance metrics such as precision, recall, and area under the receiver operating characteristic curve (AUC-ROC). These confirm the model’s sensitivity and specificity, attesting to its reliability in replicating expert-level diagnostic interpretations.
Ethical considerations in leveraging AI for pathology are also addressed, with the authors emphasizing the model’s role as a supportive tool rather than a replacement for expert judgment. This balanced perspective acknowledges the essential synergy between human expertise and machine efficiency necessary for advancing neuroscience research.
The research team envisions future iterations incorporating multi-modal data inputs, such as integrating immunohistochemical markers or transcriptional profiling results, to build even more comprehensive disease models. Combining spatial pathology with molecular signatures could open new avenues for unraveling mechanistic pathways driving synucleinopathy progression.
This impressive fusion of artificial intelligence and neuropathology stands at the forefront of a paradigm shift, heralding an era where data-driven, high-resolution disease mapping informs precision medicine strategies. The deployment of CNN-based automated histopathology presents a compelling blueprint for transformative research tools tailored to the complexities of neurological disease.
As synucleinopathies continue to challenge therapeutic development due to their heterogeneity and elusive pathology, such automated approaches provide an essential step toward unraveling these complexities. The ability to objectively and efficiently characterize pathological substrates will empower researchers to dissect the intricacies of neurodegeneration with newfound clarity.
The broader implications of this study also highlight the growing intersection of machine learning and biomedical sciences. As computational power grows and data repositories expand, the integration of AI-driven analytics is poised to accelerate discoveries across numerous domains of human health and disease.
In summary, the pioneering work by Barber-Janer and collaborators sets a new standard in histopathological analysis, bridging the gap between complex brain-wide pathological assessments and scalable, reproducible data analytics. This confluence of artificial intelligence and neuropathology not only advances our understanding of synucleinopathies but also exemplifies the transformative potential of integrating technology into biomedical research.
Subject of Research: Development of convolutional neural networks for automated brain-wide histopathological analysis in mouse models of synucleinopathies.
Article Title: Development of convolutional neural networks for automated brain-wide histopathological analysis in mouse models of synucleinopathies.
Article References:
Barber-Janer, A., Van Acker, E., Vonck, E. et al. Development of convolutional neural networks for automated brain-wide histopathological analysis in mouse models of synucleinopathies. npj Parkinsons Dis. 11, 317 (2025). https://doi.org/10.1038/s41531-025-01170-1
Image Credits: AI Generated